SUMMARY
The sam2bam is a high-throughput software tool framework that enables users to significantly speed up pre-processing for next-generation sequencing data, especially on single-node multi-core large-memory systems. The sam2bam provides file format conversion from SAM to BAM, as a basic feature. Additional features such as analyzing, filtering, and converting input data are provided by using plug-in tools, e.g., duplicate marking, which can be attached to sam2bam at runtime.
FUNCTIONALITY
| Function | Plug-in |
|---|---|
| SAM->BAM conversion | N/A |
| SAM->BAM + Alignment selection | pre_filter |
| SAM->BAM + Alignment sorting | sort_by_corrdinate |
| SAM->BAM + Duplicate marking | ibm_markdup (OpenPOWER only) |
See “HOW TO RUN” below for command lines
PERFORMANCE
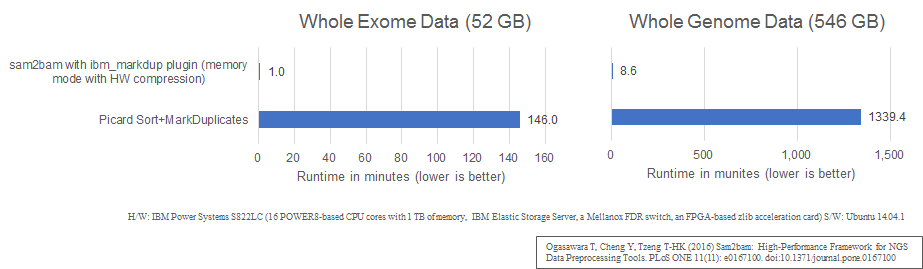
PAPER
Ogasawara T, Cheng Y, Tzeng T-HK (2016) Sam2bam: High-Performance Framework for NGS Data Preprocessing Tools. PLoS ONE 11(11): e0167100. doi:10.1371/journal.pone.0167100
HOW TO BUILD
Step 1. Download the latest build script
$ wget https://github.com/t-ogasawara/sam-to-bam/raw/master/build.sh
Step 2. Run the script on the directory where you put the source files and compile them
$ bash build.sh
HOW TO RUN
Convert the file format from SAM to BAM
$ build/samtools/samtools sam2bam -oout.bam in.sam
Preview of a new feature - processing multiple SAM files without a separate merge step (the performance will be improved in a future release)
$ build/samtools/samtools sam2bam -oout.bam in1.sam in2.sam in3.sam ...
Command-line options:
out.bam output BAM file name
in.sam input SAM file name
Run duplicate marking (OpenPOWER only)
$ build/samtools/samtools sam2bam -Fibm_markdup:r -oout.bam in.sam
Command-line options:
-Fibm_markdup: Mark duplicates
-Fibm_markdup:r Remove duplicates
-p Enable the storage mode to reduce memory footprint
(default: the memory mode)
In the storage mode, sam2bam does not keep converted BAM
records in physical memory but saves them to temporary
files. The files are created on the current directory by
default. The I/O performance of the device where the
files are created is critical for the storage mode.
BAM_PAGEFILE Change the directory where sam2bam creates temporary files
in the storage mode
The current directory is not on a fast device that you have
(e.g., mounted at /fast), BAM_PAGEFILE=/fast/bf specifies
the fast device to be used for temporary files.
Select alignments while converting the file format from SAM to BAM
$ build/samtools/samtools sam2bam -Fpre_filter:s=20:5000000-60000000 -oout.bam in.sam
$ build/samtools/samtools sam2bam -Fpre_filter:r=H06HD.2 -oout.bam in.sam
$ build/samtools/samtools sam2bam -Fpre_filter:l=Solexa-135852 -oout.bam in.sam
$ build/samtools/samtools sam2bam -Fpre_filter:q=12 -oout.bam in.sam
$ build/samtools/samtools sam2bam -Fpre_filter:f=1 -oout.bam in.sam
$ build/samtools/samtools sam2bam -Fpre_filter:F=2 -oout.bam in.sam
Command-line options:
-Fpre_filter:s=20:5000000-60000000 Select regions (e.g., including 5000000-6000000 of reference sequence 20)
-Fpre_filter:r=H06HD.2 Select read group (e.g., H06HD.2)
-Fpre_filter:l=Solexa-135852 Select library (e.g., Solexa-135852)
-Fpre_filter:q=12 Select high MAPQ (e.g., more than or equal 12)
-Fpre_filter:f=1 Select flag bits (e.g., bit 1 is set)
-Fpre_filter:F=2 Select cleared flag bits (e.g., bit 2 is not set)
Sort alignments by coordinate
$ build/samtools/samtools sam2bam -Fsort_by_coordinate: -oout.bam in.sam
Command-line options:
-Fsort_by_coordinate: sort alignments by coordinate
Use GenWQE compression accelerator (OpenPOWER only)
Prerequisites:
1. GenWQE software is installed on the system.
2. A GenWQE compression card is installed on the system.
$ export HW_ZLIB=8
$ build/samtools/samtools sam2bam -oout.bam in.sam
Command-line options:
HW_ZLIB Non-zero HW_ZLIB enables H/W acceleration of compression if the accelerator is available in the system
8 the number of threads that offload compression to the hardware accelerator